About Me
I am a Computer Science PhD candidate at Columbia University advised by David A. Knowles at the New York Genome Center. My work is supported by the NSF Graduate Research Fellowship Program (GRFP).
My research focuses on applying deep learning and generative modeling to learn single-cell dynamics and alternative splicing mechanisms in neurodegenerative disease. Utilizing AI methods to derive biological insights from multiomics is an exciting research avenue that will shape the future of health and medicine, and I look forward to continuing my exploration of this unique intersection of machine learning and biology.
- Deep Learning
- Generative Modeling
- Single Cell Dynamics
- Alternative Splicing
PhD in Computer Science, 2027
Columbia University
MS in Computer Science, 2024
Columbia University
BS in Computer Science, 2022
University of California, Los Angeles
Featured Publications
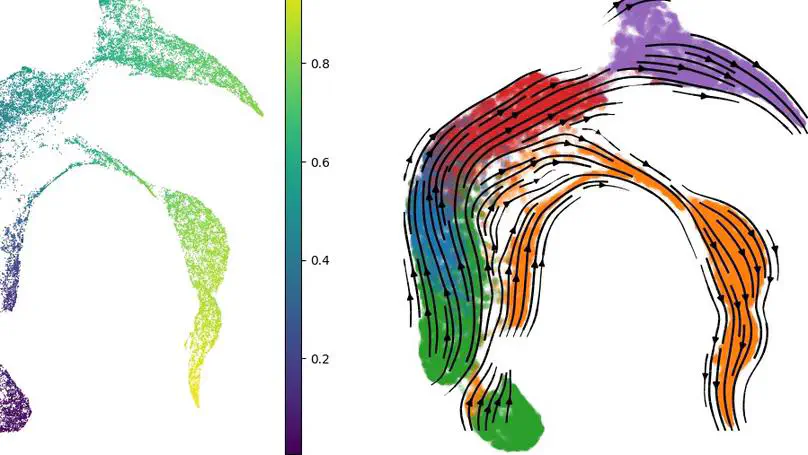
RNA velocity-based methods estimate cellular dynamics and cell developmental trajectories based on spliced and unspliced RNA counts. In this work, we introduce a new architecture, CellFlows, which incorporates self-supervised neural dimensionality reduction with the flexibility of neural-based latent time estimation into a mechanistic model, improving model interpretability and accuracy. CellFlows models splicing dynamics to infer gene and context-specific kinetic rates at single-cell resolution and correctly identifies both linear and branching cellular differentiation pathways originating from mouse embryonic stem cells.
Publications
Experience
Current projects include:
- Deep generative modeling of splicing dynamics in single cells. CellFlows presented in ICML'24 Workshop ML for Life and Material Sciences.
- Causal structure learning of gene regulatory networks from large-scale perturbations of single cells
- Deep learning-based prediction of alternative splicing from pre-mRNA sequence
- Probabilistic differential analysis of alternative splicing in neurodegenerative disease